RNA expression from almost 1,000 genes mapped at single cell and subcellular resolution within non-small cell lung cancer samples
NanoString Technologies, Inc. (NASDAQ: NSTG), a leading provider of life science tools for discovery and translational research, today announced the first public release of data generated by the CosMx™ Spatial Molecular Imager (SMI). The data represents the first high-plex, single cell in situ dataset from formalin-fixed paraffin-embedded (FFPE) tissue samples and is accompanied by the simultaneous publication of a manuscript describing the CosMx SMI technology and performance.
This press release features multimedia. View the full release here: https://www.businesswire.com/news/home/20211109005329/en/
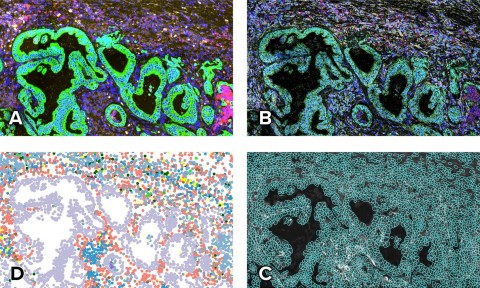
The CosMx SMI dataset contains a protein morphology map (A), a subcellular expression map of 960 genes (B), cell segmentation (C), and a spatially-resolved cell type map (D) of non-small cell lung cancer tissues. (Photo: Business Wire)
CosMx SMI enables high-resolution imaging of hundreds or thousands of RNAs and proteins within morphologically intact whole tissue sections. The technology combines the power of high-plex profiling with high-resolution imaging, allowing researchers to visualize and quantify gene expression and protein at single cell and subcellular resolutions with both fresh frozen and FFPE tissue samples.
"The ability to characterize both gene expression levels and the topography of single cells within native tissue context will yield new insights into disease pathogenesis and determinants of treatment response," said Christina Curtis, Ph.D., MSc, Associate Professor of Medicine and Genetics, Stanford University School of Medicine. “Importantly, CosMx SMI's compatibility with FFPE samples makes it a valuable platform for unlocking insights from clinical and bio-banked specimens.”
NanoString used the CosMx SMI to characterize FFPE tissue from eight different non-small cell lung cancer (NSCLC) samples from five patients. The SMI data creates a spatial cell atlas of NSCLC tissue by defining a subcellular expression map of 960 genes and a single cell map of 18 identified cell types.
The CosMx SMI dataset is open-source spatial in situ imaging and molecular data that can be accessed through NanoString’s website (www.nanostring.com/cosmx-dataset) and analyzed by the research community. The complete dataset consists of over 800,000 single cells and ~260 million transcripts, and a spatially-resolved cell type map of NSCLC tissue across a ~150 mm2 area.
The data release is accompanied by the simultaneous publication of a manuscript available on BioRxiv entitled, “High-Plex Multiomic Analysis in FFPE Tissue at Single-Cellular and Subcellular Resolution by Spatial Molecular Imaging.” The paper describes CosMx SMI’s chemistry, sensitivity, and reproducibility, with detailed discussion of the publicly released NSCLC dataset. In addition, the manuscript demonstrates the proteomic power of SMI using an 81-plex antibody panel to map protein expression within FFPE breast cancer samples.
"NanoString’s mission is to map the universe of biology," said Joseph Beechem, Ph.D., chief scientific officer, NanoString. “Together, the GeoMx DSP and the CosMx SMI platform span the continuum of spatial biology applications and will serve the needs of both discovery and translational researchers. The GeoMx DSP is the leading platform for measuring whole transcriptome in multi-cellular regions, and the CosMx SMI extends capability to single cell and subcellular resolution for targeted panels of up to one thousand targets.”
The CosMx SMI is expected to launch in the second half of 2022. In the meantime, researchers can apply the power of SMI to their tissue samples through the SMI Technology Access Program service provided by NanoString. For more information, email smitap@nanostring.com.
To learn more about NanoString’s CosMx Spatial Molecular Imager, visit www.nanostring.com/CosMx
About NanoString Technologies, Inc.
NanoString Technologies is a leading provider of life science tools for discovery and translational research. The company provides three platforms that allow researchers to map the universe of biology. The nCounter® Analysis System, cited in more than 4,900 peer-reviewed publications, offers a way to easily profile the expression of hundreds of genes, proteins, miRNAs, or copy number variations, simultaneously with high sensitivity and precision. NanoString’s GeoMx® Digital Spatial Profiler enables highly multiplexed spatial profiling of RNA and protein targets in various sample types, including FFPE tissue sections, and has been cited in more than 75 peer-reviewed publications. The CosMx Spatial Molecular Imager, with commercial availability expected in 2022, enables highly sensitive, high-resolution imaging of hundreds to thousands of RNAs or proteins directly from single cells within morphologically intact whole tissue sections. For more information, visit www.nanostring.com.
NanoString, NanoString Technologies, the NanoString logo, CosMx, GeoMx, and nCounter are trademarks or registered trademarks of NanoString Technologies, Inc. in various jurisdictions.
View source version on businesswire.com: https://www.businesswire.com/news/home/20211109005329/en/
Contacts
Doug Farrell
Vice President, Investor Relations & Corporate Communications
dfarrell@nanostring.com
Phone: 206-602-1768













